How to use LangChain and GPT to ask question on multiple pdf documents

We all know the power of GPT-3.5 and GPT-4 by now - it's incredible how versatile we can use these LLMs to be much more productive in our workspace. Nevertheless, under the hood, the GPT-models are just Language Models - meaning they are quite good with language tasks as well as by answering questions about their training data.
Wouldn't it be nice to also use these models to ask questions about other data sources like pdf files? Well, that's where LangChain enters the ring.
LangChain is a technology used to connect LLMs to real-world tools like Python, document archives, csv files or databases.
So, why not use LangChain to connect GPT to my pdf archives? This is exactly what we are going to do in this guide.
Using LangChain and GPT-3.5 or GPT-4 to ask questions about your pdf files
As discussed in this introductory post, LangChain is a set of tools to connect Large Language Models (LLMs) like GPT-3.5 and GPT-4 to various tools. Think python code, calculators, documents, the internet and many more.
They first of all provide easy-to-use connectors to all of these tools and even more importantly they have a concept called [Chains] - which basically is the interconnection of one or multiple actions and prompts involving an LLM.
Secondly, they provide easy-to-maintain "memory" or chat history to build powerful chat-bots. LLMs by default don't remember what you discussed just seconds ago. LangChain steps in and helps to manage this memory state.
Thirdly, they offer easy-to-use interfaces to many Vector Stores. For a quick introduction on what these stores are, have a look at this introduction. Why do we need them? Well, we can't ship the full pdf file to the LLM, as any LLM is limited by it's context window. For GPT-3.5 for example we can only have a conversion of 4000 Tokens (approx. 3000 words) - meaning any reasonably sized pdf file hit this limit. By storing so-called Embeddings - which are just high-dimensional, numeric representations of text - in a Vector Store, we can compare different texts and find their similarities. It turns out, embeddings, or vector representations of texts which are similar have a shorter vectorial distance than texts which are not similar. And vector stores are great in finding the distance of vectors.
How does this help? We can simply divide our pdf in chunks of smaller text-blocks, create embeddings of these chunks and then compare them with the question we want to ask the LLM about our pdf. For example, if we ask the question "What is photosynthesis?" we can compare the vector representation of this question with all the vectors of the text chunks of our pdf file. If there are some chunks which are similar to this question, they are highly likely to answer it. As a next step, we only send the similar chunks of text plus the question to LLM - drastically reducing the required context.
All these LangChain-tools allow us to build the following process:
- We load our pdf files and create embeddings - the vectors described above - and store them in a local file-based vector database
- We use vector similarity search to find the chunks needed to answer our question
- We send these chunks and the question to GPT-3.5 and GPT-4
- We add additional Chat capabilities to not only ask simple questions, but engage in a conversation - including conversation history
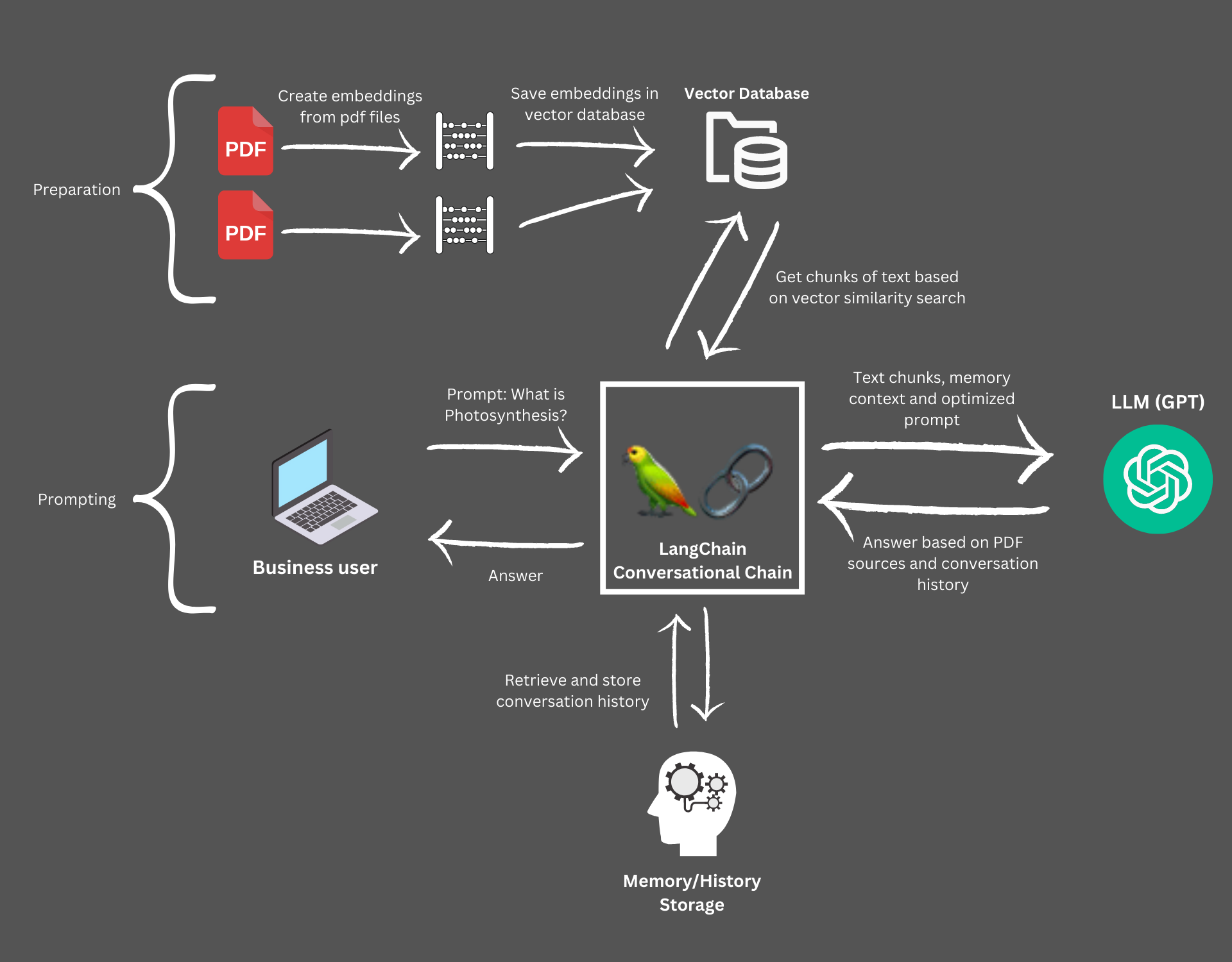 LangChain to connect PDFs to GPT-4
LangChain to connect PDFs to GPT-4
Step by step guide for using LangChain and GPT to ask questions about your pdf files
With our basic architecture established let's get our hands dirty.
-
Install the dependency modules
1pip install langchain openai pypdf chroma -
Download the sample pdf files from ResearchGate and USGS
-
Import the required modules
1from langchain.document_loaders import PyPDFLoader2from langchain.vectorstores import Chroma3from langchain.chat_models import ChatOpenAI4from langchain.embeddings.openai import OpenAIEmbeddings5from langchain.text_splitter import RecursiveCharacterTextSplitter6from langchain.chains import RetrievalQA, ConversationalRetrievalChain7import os -
Get an OpenAI key from the OpenAI platform
1os.environ["OPENAI_API_KEY"] = "sk-secretxxxxx" -
Load a pdf file, using the LangChain pypdf loader
1loader = PyPDFLoader("sample_data/Photosynthesis.pdf") -
Split the text in chunks, using one of the LangChain Text Splitters. We'll use the
RecursiveCharacterTextSplitteras this splitters tries to not split paragraphs - meaning it varies the length of the chunks, when it can keep paragraphs together.1text_splitter = RecursiveCharacterTextSplitter(chunk_size=1000, chunk_overlap=200)2pages = loader.load_and_split(text_splitter) -
Create a persistent, file-based vector store. We create a Chroma vector store. It provides good performance and is very easy to manage. It persists the vectors to the file system. Please note to set the
persist_directoryparameter - otherwise your store will only be in-memory.1directory = 'index_store'2vector_index = Chroma.from_documents(pages, OpenAIEmbeddings(), persist_directory=directory)3vector_index.persist() # actually the Chroma client automatically persists the indexes when it is disposed - however better save then sorry :-) -
Create the retriever and the query-interface. The retriever is used to get similar vectors from the vector store and the query-interface to then ask our questions. The parameter
kin the retriever sets the amount of text-chunks we want to retrieve from the vector store.1retriever = vector_index.as_retriever(search_type="similarity", search_kwargs={"k":6})2# create the chain for allowing us to chat with the document3qa_interface = RetrievalQA.from_chain_type(llm=ChatOpenAI(), chain_type="stuff", retriever=retriever, return_source_documents=True) -
Now we are ready to query our pdf files! First, let's ask about
aerobic respiration- a term described in our Photosynthesis pdf file1qa_interface("What is aerobic respiration? Return 3 paragraphs and a headline as markdown.")Result (shortened):
1{'query': 'What is aerobic respiration? Return 3 paragraphs and a headline as markdown.',2'result': '# Aerobic Respiration\n\nUnlike photosynthesis, aerobic respiration is an exergonic process (negative /Delta1G◦) with the energy released being used by the organism to power biosynthetic processes that allow growth and renewal, mechanical work (such as muscle contraction or flagella rotation) and facilitating changes in chemical concentrations within the cell (e.g. accumulation of nutrients and expulsion of waste). The use of exergonic reactions to power endergonic ones associated with biosynthesis and housekeeping in biological organisms such that the overall free energy change is negative is known as ‘coupling’.\n\nPhotosynthesis and respiration are thus seemingly the reverse of one another, with the important caveat that both oxygen formation during photosynthesis and its utilization during respiration result in its liberation or incorporation respectively into water rather than CO2. In addition, glucose is one of several possible products of photosynthesis with amino acids and lipids also being synthesized rapidly from the primary photosynthetic products.\n\nThe inefficiencies of the Rubisco enzyme mean that plants must produce it in very large amounts ( ∼30–50% of total soluble protein in a spinach leaf) to achieve the maximal photosynthetic rate.',3'source_documents': [Document(page_content='photosynthesis acts as the ... is negative is', metadata={'source': 'sample_data/Photosynthesis.pdf', 'page': 1}),4Document(page_content='Essays in Biochemistry (2016) 60255–273\nDOI: 10.1042/EBC20160016\nFigure 1. The glob...an', metadata={'source': 'sample_data/Photosynthesis.pdf', 'page': 2}),5Document(page_content='a st h eb a s i sf o r...', metadata={'source': 'sample_data/Photosynthesis.pdf', 'page': 0}),6Document(page_content='during respiration, decay....', metadata={'source': 'sample_data/Photosynthesis.pdf', 'page': 1}),7Document(page_content='with biosynthesis ....', metadata={'source': 'sample_data/Photosynthesis.pdf', 'page': 1}),8Document(page_content='reaction, ...4.0 (CC BY).271', metadata={'source': 'sample_data/Photosynthesis.pdf', 'page': 16})]}
Look at that! The question is perfectly answered! Not only do we get the answer but also which documents were used as source files. Especially for more serious endeavours, this is very convenient. If you don't want the source documents, set the return_source_documents parameter in the qa_interface to False.
Adding additional documents to our vector store
So, now that we know how to ask questions about one specific document, let's add additional documents. These can be totally different documents about different topics. The vector similarity search will yield the correct documents when asking our questions. This is an awesome way for building a knowledge base. Just index any document you want to have as source data available and ask questions about any one of them.
1loader = PyPDFLoader("sample_data/graphite.pdf")23pages_new = loader.load_and_split(text_splitter)45_ = vector_index.add_documents(pages_new)6vector_index.persist()
As you might notice, the process is more or less the same as for the first document:
- Load the document
- Split the document, using the same text splitter as before
- Add the document to our existing index
That's all the magic - now we can ask questions to either one of the documents.
Adding memory to our conversations
Up until now we used a RetrievalQA chain - which is a LangChain chain type specifically tailored for loading chunks of documents from a vector store and asking a single question about them.
However, sometimes it would be convenient to really have a full conversation about a document and being able to refer to already discussed things.
For such a thing to work, our system needs memory or a conversation history. Conveniently, LangChain got as covered also here. Instead of the RetrievalQA - chain we'll use the ConversationalRetrievalChain.
1conv_interface = ConversationalRetrievalChain.from_llm(ChatOpenAI(temperature=0), retriever=retriever)
Next, we initialize our chat history - this might be a simple list.
1chat_history = []2query = "What is photosyntheses?"
And that's all the preparation we need. Let's start our chat conversation.
1result = conv_interface({"question": query, "chat_history": chat_history})2result["answer"]
Result: 'Photosynthesis is the process of converting water and CO2 into complex organic molecules such as carbohydrates and oxygen. It involves two reactions, the light reactions and the dark reactions, which occur in separate parts of the chloroplast. The light reactions involve the splitting of water into oxygen, protons and electrons, while the dark reactions involve the reduction of CO2 to carbohydrate. Photosynthesis is the primary energy input into the global food chain and is essential for the formation of the ozone layer, the evolution of aerobic respiration and complex multicellular life.'
Ok, this is nice - but a little too long, right? As we have a chat interface - let's simply ask to shorten this sentence. Before that however, let's not forget to add the previous conversation to our chat history.
1# Add previous conversation to chat history2chat_history.append((query, result["answer"]))34# Query to shorten the last sentence5query = "Can you shorten this sentence please?"6result = conv_interface({"question": query, "chat_history": chat_history})7result["answer"]
Result: 'Photosynthesis is the process by which plants use light energy to convert water and carbon dioxide into oxygen and energy-rich organic compounds. This process sustains virtually all life on Earth, providing oxygen and food, and forms the basis of global food chains.
Voila, here you have it. Without specifying which sentence to shorten, LangChain was able to automatically extract this information from the chat history. From here, we could simply continue our conversation, until we hit the context limit of the LLM.
What happens under the hood`
The ConversationalRetrievalChain provides a very powerful interface, as it allows to seamlessly add historical context or memory to our chain. Now, under the hood, LangChain executes two prompts and a vector store retrieval:
- The first prompt is used to summarize the whole memory as well as the new query into a single question.
- Then, this new question is used to find the most relevant piece of information in the vector store
- Finally, the relevant chunks of information as well as the refined questions are sent to the LLM for the final answer
This is great because it not only smartly combines history and new questions - but it also drastically shortens the required context, as you don't always need to send the full history to the LLM.
Loading persisted embeddings
The complete this guide, let's also have a look at how to load previously persisted vector data/embeddings from disk.
1vector_index = Chroma(persist_directory=directory, embedding_function=OpenAIEmbeddings())
That's all we need - now you can use this index in the retriever as before.
Summary
In this blog post, we demonstrated how to use LangChain to connect multiple PDF files to GPT-3.5 and GPT-4 and engage in a conversation about these files. Through LangChain, users can leverage GPT models to ask questions about other data sources like PDFs.
The post provided a step-by-step guide on using LangChain and GPT to ask questions about PDF files. It began by installing the necessary dependencies, then moved on to loading a sample PDF file and splitting its text into smaller chunks for processing. We then created a local file-based vector store to save the PDF's embeddings and built a retriever and a query interface to find relevant text blocks and ask questions to GPT models.
By following the guide, users can query their PDF files, add additional documents to their vector stores, and engage in a conversation with their data by keeping track of chat history. The post also covered loading previously persisted vector data from disk.
The result is a powerful and flexible way to interact with data from multiple sources, perfect for building a knowledge base or working with diverse documents.
For reference, find here the full code example:
1from langchain.document_loaders import PyPDFLoader2from langchain.vectorstores import Chroma3from langchain.chat_models import ChatOpenAI4from langchain.embeddings.openai import OpenAIEmbeddings5from langchain.text_splitter import RecursiveCharacterTextSplitter6from langchain.chains import RetrievalQA, ConversationalRetrievalChain7import os89# OpenAI platform key10os.environ["OPENAI_API_KEY"] = "sk-secretxxxxx"1112# Load pdf file and split into chunks13loader = PyPDFLoader("sample_data/Photosynthesis.pdf")14text_splitter = RecursiveCharacterTextSplitter(chunk_size=1000, chunk_overlap=200)15pages = loader.load_and_split(text_splitter)1617# Prepare vector store18directory = 'index_store'19vector_index = Chroma.from_documents(pages, OpenAIEmbeddings(), persist_directory=directory)20vector_index.persist() # actually the Chroma client automatically persists the indexes when it is disposed - however better save then sorry :-)2122# Prepare the retriever chain23retriever = vector_index.as_retriever(search_type="similarity", search_kwargs={"k":6})24qa_interface = RetrievalQA.from_chain_type(llm=ChatOpenAI(), chain_type="stuff", retriever=retriever, return_source_documents=True)2526# First query27print(qa_interface("What is aerobic respiration? Return 3 paragraphs and a headline as markdown."))2829#Adding additional docs30loader = PyPDFLoader("sample_data/graphite.pdf")3132pages_new = loader.load_and_split(text_splitter)3334_ = vector_index.add_documents(pages_new)35vector_index.persist()3637#Adding memory38conv_interface = ConversationalRetrievalChain.from_llm(ChatOpenAI(temperature=0), retriever=retriever)3940chat_history = []41query = "What is photosyntheses?"4243# First chat query44result = conv_interface({"question": query, "chat_history": chat_history})45print(result["answer"])4647# Second query, using the previous queries as memory48# Add previous conversation to chat history49chat_history.append((query, result["answer"]))5051# Shorten the last sentence52query = "Can you shorten this sentence please?"53result = conv_interface({"question": query, "chat_history": chat_history})54print(result["answer"])
What's next?
If you want to analyze csv files and even create plots from your csvs, have a look at this next post in this LangChain blogging series.
If you want to "chat" with your BigQuery data, have a look at this tutorial, detailing how to connect GPT to BigQuery.
------------------
Interested in how to train your very own Large Language Model?
We prepared a well-researched guide for how to use the latest advancements in Open Source technology to fine-tune your own LLM. This has many advantages like:
- Cost control
- Data privacy
- Excellent performance - adjusted specifically for your intended use